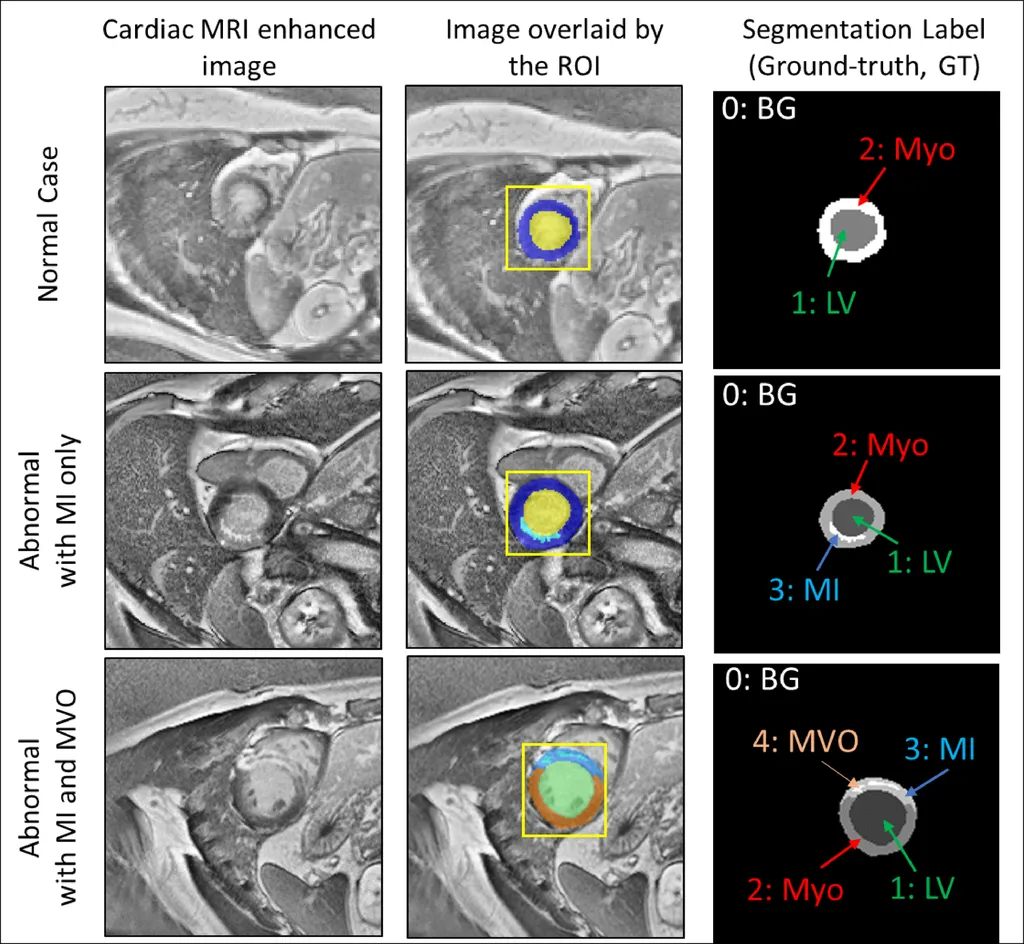
In the world of medical imaging, few-shot learning (FSL) has been a game-changer, especially in cardiac MRI segmentation. The challenge, however, has been the reliance on semi-supervised techniques that are sensitive to domain shifts and validation bias, which limits the ability to generalize to unseen data. Enter PathCo-LatticE, a fully supervised FSL framework that’s shaking things up.
The innovation behind PathCo-LatticE is the replacement of unlabeled data with pathology-guided synthetic supervision. The first step in this process is the Virtual Patient Engine, which models continuous latent disease trajectories from sparse clinical anchors. This is done using generative modeling to synthesize physiologically plausible, fully labeled 3D cohorts. In simpler terms, it’s like creating a virtual library of cardiac MRI images that represent a wide range of diseases and severities, all fully labeled and ready for training.
The second part of the framework is Self-Reinforcing Interleaved Validation (SIV). This provides a leakage-free protocol that evaluates models online with progressively challenging synthetic samples. The result? No need for real validation data, which is a significant step forward in reducing bias and improving model performance.
Finally, a dynamic Lattice-of-Experts (LoE) organizes specialized networks within a pathology-aware topology. This means that the most relevant experts (or networks) are activated per input, enabling robust zero-shot generalization to unseen data without target-domain fine-tuning. It’s like having a team of specialists, each an expert in a specific area, ready to tackle any challenge that comes their way.
The researchers evaluated PathCo-LatticE in a strict out-of-distribution (OOD) setting, deriving all anchors and severity statistics from a single-source domain (ACDC) and performing zero-shot testing on the multi-center, multi-vendor M&Ms dataset. The results were impressive. PathCo-LatticE outperformed four state-of-the-art FSL methods by 4.2-11% Dice starting from only 7 labeled anchors, and approached fully supervised performance (within 1% Dice) with only 19 labeled anchors. The method also showed superior harmonization across four vendors and generalization to unseen pathologies.
The implications of this research are significant. By mitigating data scarcity and reducing bias, PathCo-LatticE could greatly improve the accuracy and reliability of cardiac MRI segmentation. This, in turn, could lead to better diagnosis and treatment of cardiac diseases. The code for PathCo-LatticE will be made publicly available, opening up new avenues for research and development in the field.